CRAN dependencies
New policy in CRAN
With the new policy of a maximum of 20 packages in Imports. Let’s see how many dependencies has each package on CRAN and Bioconductor:
CRAN
ap <- available.packages()
dp <- tools::package_dependencies(rownames(ap), db = ap, which = "Imports",
recursive = FALSE)
dp_n <- lengths(dp)
tb_dp <- sort(table(dp_n), decreasing = TRUE)
barplot(tb_dp)
Most of the packages has 0 dependencies and just 212 has 20 dependencies or more:
names(dp_n)[dp_n >= 20]
## [1] "AdhereRViz" "AFM" "AirSensor"
## [4] "alookr" "amt" "animaltracker"
## [7] "antaresViz" "BAMBI" "BasketballAnalyzeR"
## [10] "BAwiR" "bea.R" "BETS"
## [13] "bibliometrix" "biomod2" "bioRad"
## [16] "BIRDS" "bootnet" "bpcs"
## [19] "breathtestcore" "brms" "card"
## [22] "chemmodlab" "chillR" "Clustering"
## [25] "CNVScope" "codebook" "cSEM"
## [28] "ctmm" "ctsem" "DAMisc"
## [31] "dartR" "datacleanr" "dccvalidator"
## [34] "devtools" "dextergui" "diceR"
## [37] "dipsaus" "DIscBIO" "distill"
## [40] "dlookr" "dragon" "drhur"
## [43] "dyngen" "dynwrap" "ebirdst"
## [46] "ecd" "ecochange" "EcoGenetics"
## [49] "ecospat" "EFAtools" "eiCompare"
## [52] "elementR" "emdi" "emuR"
## [55] "eph" "EpiNow2" "epitweetr"
## [58] "fdm2id" "FedData" "finalfit"
## [61] "forestmangr" "genBaRcode" "geoviz"
## [64] "ggquickeda" "GJRM" "GmAMisc"
## [67] "golem" "graph4lg" "GWSDAT"
## [70] "hdpGLM" "highcharter" "hmi"
## [73] "htsr" "hybridEnsemble" "iCellR"
## [76] "immunarch" "inlmisc" "IntClust"
## [79] "iNZightTools" "IOHanalyzer" "isoreader"
## [82] "ITNr" "jmv" "jsmodule"
## [85] "JWileymisc" "KarsTS" "lilikoi"
## [88] "mdapack" "memapp" "metacoder"
## [91] "MetaDBparse" "MetaIntegrator" "microbial"
## [94] "missCompare" "mlflow" "modchart"
## [97] "modeltime" "modeltime.ensemble" "modeltime.resample"
## [100] "momentuHMM"
## [ reached getOption("max.print") -- omitted 112 entries ]Bioconductor
The interesting part is discovering how to add the repository.
The trick is to make us of BiocManager::repositories():
BioC_software <- BiocManager::repositories()["BioCsoft"]
bp <- available.packages(contriburl = contrib.url(BioC_software))
dp_BioC <- tools::package_dependencies(rownames(bp), db = bp, which = "Imports",
recursive = FALSE)
dp_BioC_n <- lengths(dp_BioC)
tb_dp_BioC <- sort(table(dp_BioC_n), decreasing = TRUE)
barplot(tb_dp_BioC)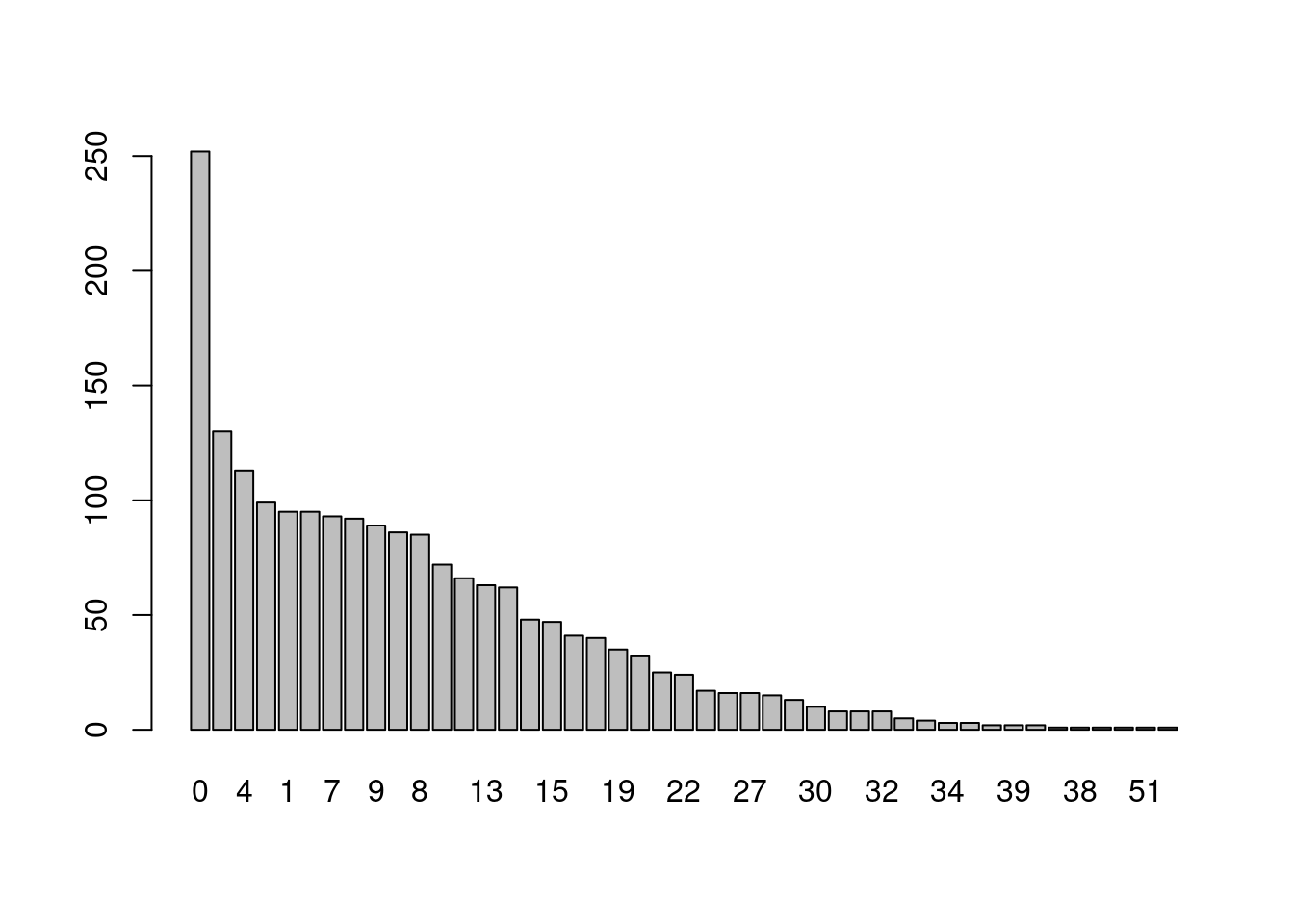
It seems that software packages on Bioconductor tend to have more dependencies than on CRAN. If this policy would be implemented on Bioconductor it would affect 219 packages:
names(dp_BioC_n)[dp_BioC_n >= 20]
## [1] "abseqR" "adductomicsR" "ALPS"
## [4] "AlpsNMR" "AMARETTO" "amplican"
## [7] "AneuFinder" "animalcules" "appreci8R"
## [10] "ArrayExpressHTS" "arrayQualityMetrics" "artMS"
## [13] "ASpediaFI" "ATACseqQC" "BASiCS"
## [16] "BatchQC" "bigPint" "bioCancer"
## [19] "BiocOncoTK" "BiocPkgTools" "biovizBase"
## [22] "biscuiteer" "BPRMeth" "bsseq"
## [25] "BUSpaRse" "CAGEr" "CATALYST"
## [28] "celda" "CEMiTool" "CeTF"
## [31] "ChAMP" "chimeraviz" "chipenrich"
## [34] "ChIPpeakAnno" "ChIPQC" "ChIPseeker"
## [37] "ChromSCape" "chromVAR" "cicero"
## [40] "circRNAprofiler" "CiteFuse" "clusterExperiment"
## [43] "clustifyr" "CNEr" "CNVPanelizer"
## [46] "CNVRanger" "cola" "COMPASS"
## [49] "compcodeR" "CONFESS" "consensusDE"
## [52] "contiBAIT" "crlmm" "crossmeta"
## [55] "cTRAP" "CytoML" "CytoTree"
## [58] "DAMEfinder" "DaMiRseq" "debrowser"
## [61] "deco" "decompTumor2Sig" "DEGreport"
## [64] "DEP" "DepecheR" "destiny"
## [67] "DEsubs" "DiffBind" "diffcyt"
## [70] "diffHic" "diffloop" "DiscoRhythm"
## [73] "dmrseq" "Doscheda" "EGSEA"
## [76] "ELMER" "ENmix" "enrichTF"
## [79] "esATAC" "EventPointer" "exomePeak2"
## [82] "fcoex" "FindMyFriends" "flowSpy"
## [85] "flowWorkspace" "FRASER" "GAPGOM"
## [88] "GENESIS" "GeneTonic" "genomation"
## [91] "GenomicInteractions" "GenVisR" "ggbio"
## [94] "GGtools" "GladiaTOX" "GmicR"
## [97] "gQTLstats" "Gviz" "GWENA"
## [100] "HiCBricks"
## [ reached getOption("max.print") -- omitted 119 entries ]Reproducibility
## ─ Session info ───────────────────────────────────────────────────────────────────────────────────────────────────────
## setting value
## version R version 4.0.1 (2020-06-06)
## os Ubuntu 20.04.1 LTS
## system x86_64, linux-gnu
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz Europe/Madrid
## date 2021-01-08
##
## ─ Packages ───────────────────────────────────────────────────────────────────────────────────────────────────────────
## package * version date lib source
## assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.0.1)
## BiocManager 1.30.10 2019-11-16 [1] CRAN (R 4.0.1)
## blogdown 0.21.84 2021-01-07 [1] Github (rstudio/blogdown@c4fbb58)
## bookdown 0.21 2020-10-13 [1] CRAN (R 4.0.1)
## cli 2.2.0 2020-11-20 [1] CRAN (R 4.0.1)
## crayon 1.3.4 2017-09-16 [1] CRAN (R 4.0.1)
## digest 0.6.27 2020-10-24 [1] CRAN (R 4.0.1)
## evaluate 0.14 2019-05-28 [1] CRAN (R 4.0.1)
## fansi 0.4.1 2020-01-08 [1] CRAN (R 4.0.1)
## glue 1.4.2 2020-08-27 [1] CRAN (R 4.0.1)
## htmltools 0.5.0 2020-06-16 [1] CRAN (R 4.0.1)
## knitr 1.30 2020-09-22 [1] CRAN (R 4.0.1)
## magrittr 2.0.1 2020-11-17 [1] CRAN (R 4.0.1)
## rlang 0.4.10 2020-12-30 [1] CRAN (R 4.0.1)
## rmarkdown 2.6 2020-12-14 [1] CRAN (R 4.0.1)
## sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 4.0.1)
## stringi 1.5.3 2020-09-09 [1] CRAN (R 4.0.1)
## stringr 1.4.0 2019-02-10 [1] CRAN (R 4.0.1)
## withr 2.3.0 2020-09-22 [1] CRAN (R 4.0.1)
## xfun 0.20 2021-01-06 [1] CRAN (R 4.0.1)
## yaml 2.2.1 2020-02-01 [1] CRAN (R 4.0.1)
##
## [1] /home/lluis/bin/R/4.0.1/lib/R/library