PCA and MDS
Many has been said about the relation of PCA and MDS. For instance this, or this other one on the practical point of view, however it has been different. In R we have on base (really stats package) three functions: prcomp, princomp and cmdscale
We will compare the output of the three of them on this dataset:
set.seed(547913)
x <- runif(100)
M <- matrix(x, ncol = 10, nrow = 10)prcomp
stats:::prcomp.default## function (x, retx = TRUE, center = TRUE, scale. = FALSE, tol = NULL,
## rank. = NULL, ...)
## {
## chkDots(...)
## x <- as.matrix(x)
## x <- scale(x, center = center, scale = scale.)
## cen <- attr(x, "scaled:center")
## sc <- attr(x, "scaled:scale")
## if (any(sc == 0))
## stop("cannot rescale a constant/zero column to unit variance")
## n <- nrow(x)
## p <- ncol(x)
## k <- if (!is.null(rank.)) {
## stopifnot(length(rank.) == 1, is.finite(rank.), as.integer(rank.) >
## 0)
## min(as.integer(rank.), n, p)
## }
## else min(n, p)
## s <- svd(x, nu = 0, nv = k)
## j <- seq_len(k)
## s$d <- s$d/sqrt(max(1, n - 1))
## if (!is.null(tol)) {
## rank <- sum(s$d > (s$d[1L] * tol))
## if (rank < k) {
## j <- seq_len(k <- rank)
## s$v <- s$v[, j, drop = FALSE]
## }
## }
## dimnames(s$v) <- list(colnames(x), paste0("PC", j))
## r <- list(sdev = s$d, rotation = s$v, center = if (is.null(cen)) FALSE else cen,
## scale = if (is.null(sc)) FALSE else sc)
## if (retx)
## r$x <- x %*% s$v
## class(r) <- "prcomp"
## r
## }
## <bytecode: 0x55f3a2a572b8>
## <environment: namespace:stats>M_prcomp <- prcomp(M)
plot(M_prcomp$rotation)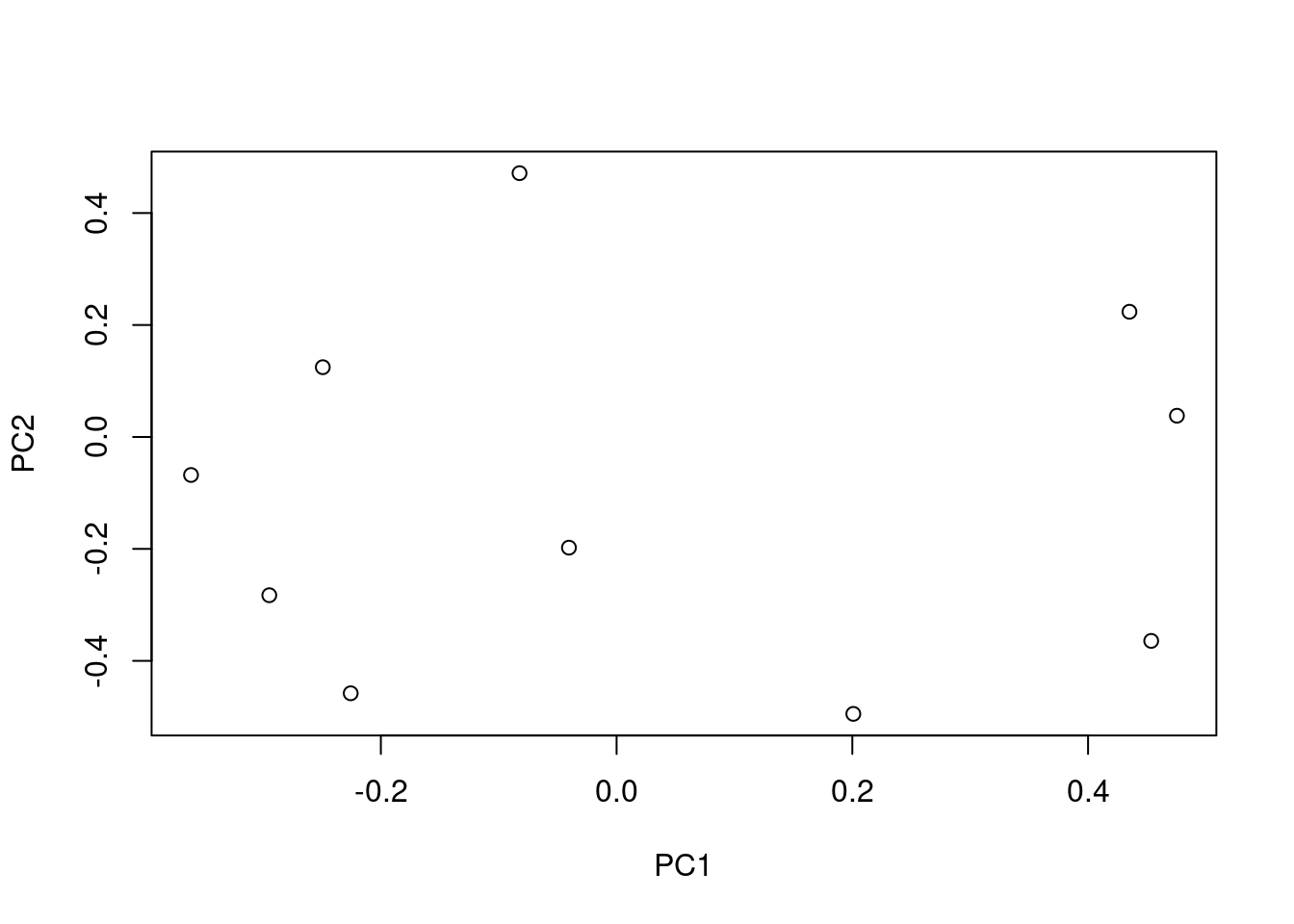
M_prcomp$rotation[1:10, 1:2]## PC1 PC2
## [1,] -0.22557903 -0.45794036
## [2,] 0.20084830 -0.49472788
## [3,] -0.36106196 -0.06796242
## [4,] -0.04042137 -0.19776900
## [5,] 0.47538969 0.03790801
## [6,] -0.29464293 -0.28278067
## [7,] -0.24926025 0.12461926
## [8,] -0.08235154 0.47127438
## [9,] 0.45357250 -0.36434371
## [10,] 0.43511396 0.22366429princomp
stats:::princomp.default## function (x, cor = FALSE, scores = TRUE, covmat = NULL, subset = rep_len(TRUE,
## nrow(as.matrix(x))), fix_sign = TRUE, ...)
## {
## chkDots(...)
## cl <- match.call()
## cl[[1L]] <- as.name("princomp")
## z <- if (!missing(x))
## as.matrix(x)[subset, , drop = FALSE]
## if (is.list(covmat)) {
## if (any(is.na(match(c("cov", "n.obs"), names(covmat)))))
## stop("'covmat' is not a valid covariance list")
## cv <- covmat$cov
## n.obs <- covmat$n.obs
## cen <- covmat$center
## }
## else if (is.matrix(covmat)) {
## if (!missing(x))
## warning("both 'x' and 'covmat' were supplied: 'x' will be ignored")
## cv <- covmat
## n.obs <- NA
## cen <- NULL
## }
## else if (is.null(covmat)) {
## dn <- dim(z)
## if (dn[1L] < dn[2L])
## stop("'princomp' can only be used with more units than variables")
## covmat <- cov.wt(z)
## n.obs <- covmat$n.obs
## cv <- covmat$cov * (1 - 1/n.obs)
## cen <- covmat$center
## }
## else stop("'covmat' is of unknown type")
## if (!is.numeric(cv))
## stop("PCA applies only to numerical variables")
## if (cor) {
## sds <- sqrt(diag(cv))
## if (any(sds == 0))
## stop("cannot use 'cor = TRUE' with a constant variable")
## cv <- cv/(sds %o% sds)
## }
## edc <- eigen(cv, symmetric = TRUE)
## ev <- edc$values
## if (any(neg <- ev < 0)) {
## if (any(ev[neg] < -9 * .Machine$double.eps * ev[1L]))
## stop("covariance matrix is not non-negative definite")
## else ev[neg] <- 0
## }
## cn <- paste0("Comp.", 1L:ncol(cv))
## names(ev) <- cn
## dimnames(edc$vectors) <- if (missing(x))
## list(dimnames(cv)[[2L]], cn)
## else list(dimnames(x)[[2L]], cn)
## sdev <- sqrt(ev)
## sc <- setNames(if (cor)
## sds
## else rep.int(1, ncol(cv)), colnames(cv))
## fix <- if (fix_sign)
## function(A) {
## mysign <- function(x) ifelse(x < 0, -1, 1)
## A[] <- apply(A, 2L, function(x) x * mysign(x[1L]))
## A
## }
## else identity
## ev <- fix(edc$vectors)
## scr <- if (scores && !missing(x) && !is.null(cen))
## scale(z, center = cen, scale = sc) %*% ev
## if (is.null(cen))
## cen <- rep(NA_real_, nrow(cv))
## edc <- list(sdev = sdev, loadings = structure(ev, class = "loadings"),
## center = cen, scale = sc, n.obs = n.obs, scores = scr,
## call = cl)
## class(edc) <- "princomp"
## edc
## }
## <bytecode: 0x55f3a466ee08>
## <environment: namespace:stats>M_princomp <- princomp(M)
plot(M_princomp$scores)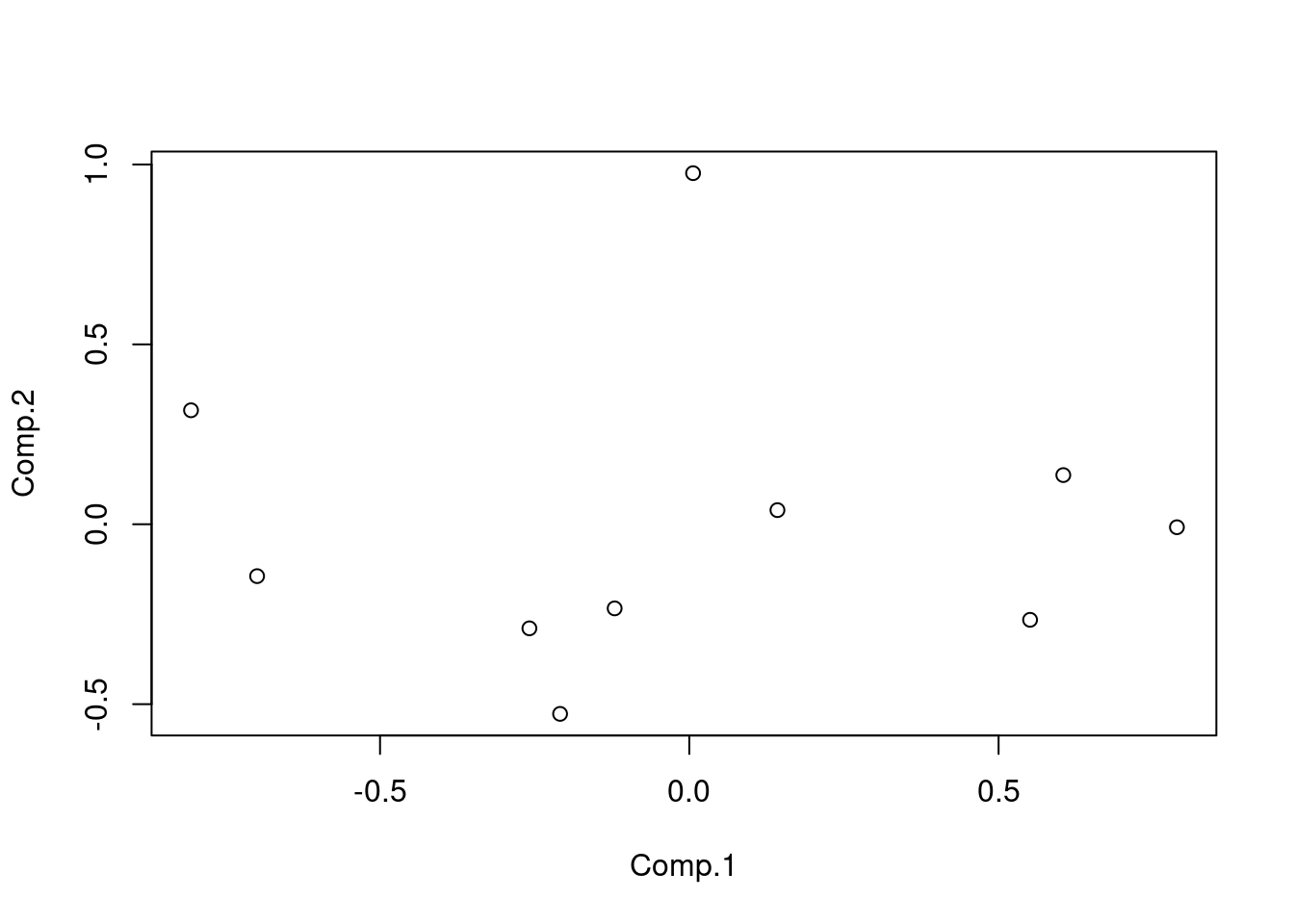
M_princomp$scores[1:10, 1:2]## Comp.1 Comp.2
## [1,] -0.805589530 0.316761323
## [2,] -0.258524683 -0.289328086
## [3,] -0.208908743 -0.527063309
## [4,] 0.142497647 0.039243307
## [5,] -0.698753661 -0.144250131
## [6,] 0.006096989 0.975808798
## [7,] 0.788321400 -0.008304383
## [8,] -0.120707570 -0.233847577
## [9,] 0.604636715 0.136609136
## [10,] 0.550931437 -0.265629080cmdscale
cmdscale## function (d, k = 2, eig = FALSE, add = FALSE, x.ret = FALSE,
## list. = eig || add || x.ret)
## {
## if (anyNA(d))
## stop("NA values not allowed in 'd'")
## if (!list.) {
## if (eig)
## warning("eig=TRUE is disregarded when list.=FALSE")
## if (x.ret)
## warning("x.ret=TRUE is disregarded when list.=FALSE")
## }
## if (is.null(n <- attr(d, "Size"))) {
## if (add)
## d <- as.matrix(d)
## x <- as.matrix(d^2)
## storage.mode(x) <- "double"
## if ((n <- nrow(x)) != ncol(x))
## stop("distances must be result of 'dist' or a square matrix")
## rn <- rownames(x)
## }
## else {
## rn <- attr(d, "Labels")
## x <- matrix(0, n, n)
## if (add)
## d0 <- x
## x[row(x) > col(x)] <- d^2
## x <- x + t(x)
## if (add) {
## d0[row(x) > col(x)] <- d
## d <- d0 + t(d0)
## }
## }
## n <- as.integer(n)
## if (is.na(n) || n > 46340)
## stop(gettextf("invalid value of %s", "'n'"), domain = NA)
## if ((k <- as.integer(k)) > n - 1 || k < 1)
## stop("'k' must be in {1, 2, .. n - 1}")
## x <- .Call(C_DoubleCentre, x)
## if (add) {
## i2 <- n + (i <- 1L:n)
## Z <- matrix(0, 2L * n, 2L * n)
## Z[cbind(i2, i)] <- -1
## Z[i, i2] <- -x
## Z[i2, i2] <- .Call(C_DoubleCentre, 2 * d)
## e <- eigen(Z, symmetric = FALSE, only.values = TRUE)$values
## add.c <- max(Re(e))
## x <- matrix(double(n * n), n, n)
## non.diag <- row(d) != col(d)
## x[non.diag] <- (d[non.diag] + add.c)^2
## x <- .Call(C_DoubleCentre, x)
## }
## e <- eigen(-x/2, symmetric = TRUE)
## ev <- e$values[seq_len(k)]
## evec <- e$vectors[, seq_len(k), drop = FALSE]
## k1 <- sum(ev > 0)
## if (k1 < k) {
## warning(gettextf("only %d of the first %d eigenvalues are > 0",
## k1, k), domain = NA)
## evec <- evec[, ev > 0, drop = FALSE]
## ev <- ev[ev > 0]
## }
## points <- evec * rep(sqrt(ev), each = n)
## dimnames(points) <- list(rn, NULL)
## if (list.) {
## evalus <- e$values
## list(points = points, eig = if (eig) evalus, x = if (x.ret) x,
## ac = if (add) add.c else 0, GOF = sum(ev)/c(sum(abs(evalus)),
## sum(pmax(evalus, 0))))
## }
## else points
## }
## <bytecode: 0x55f3a4df5778>
## <environment: namespace:stats>M_dist <- as.dist(M)
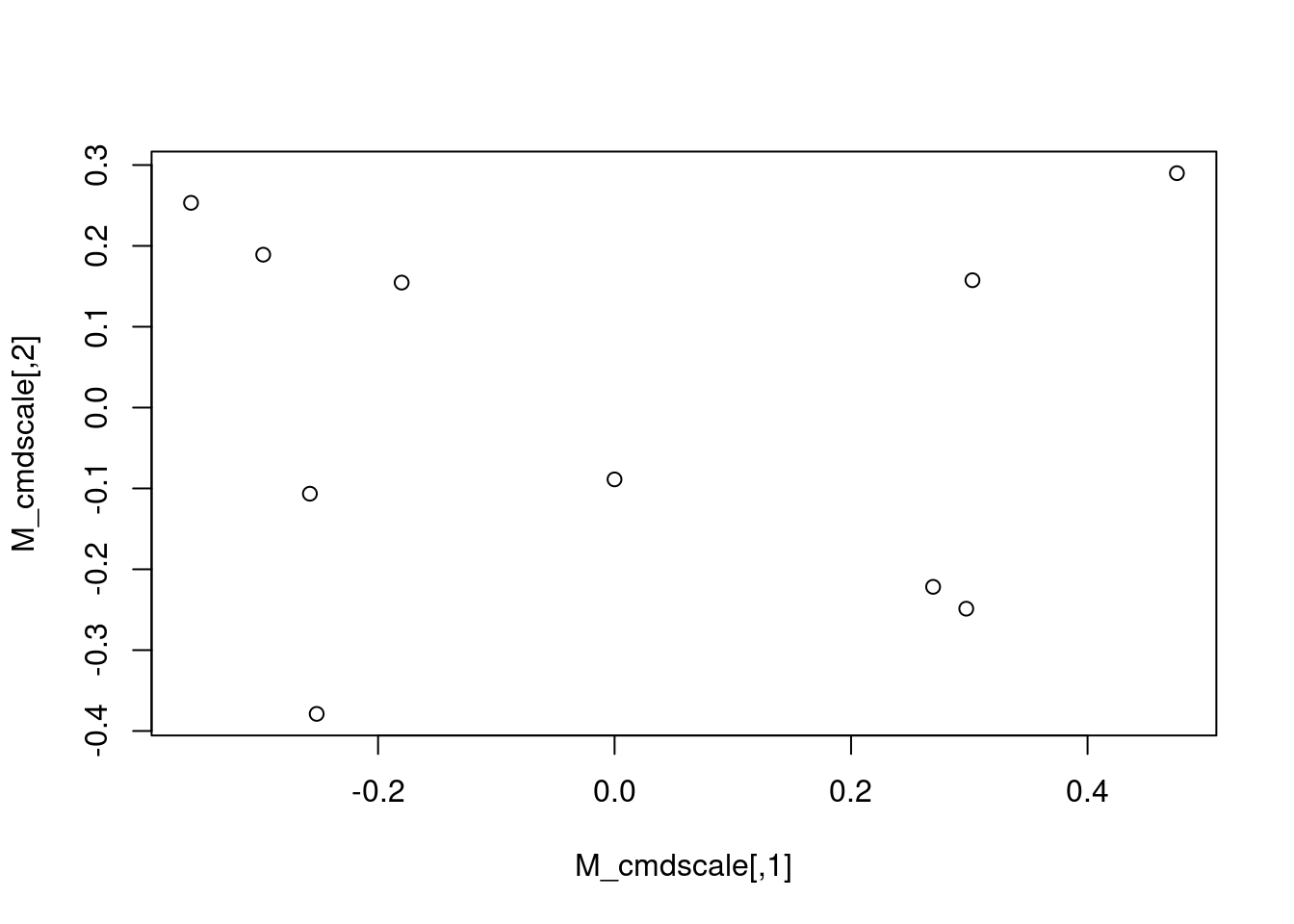
M_cmdscale <- cmdscale(M_dist)
plot(M_cmdscale)